Slopes of column-wise linear regressions of each column of a Filebacked Big Matrix, with some other associated statistics. Covariates can be added to correct for confounders.
big_univLinReg(
X,
y.train,
ind.train = rows_along(X),
ind.col = cols_along(X),
covar.train = NULL,
thr.eigval = 1e-04,
ncores = 1
)Arguments
- X
An object of class FBM.
- y.train
Vector of responses, corresponding to
ind.train.- ind.train
An optional vector of the row indices that are used, for the training part. If not specified, all rows are used. Don't use negative indices.
- ind.col
An optional vector of the column indices that are used. If not specified, all columns are used. Don't use negative indices.
- covar.train
Matrix of covariables to be added in each model to correct for confounders (e.g. the scores of PCA), corresponding to
ind.train. Default isNULLand corresponds to only adding an intercept to each model. You can usecovar_from_df()to convert from a data frame.- thr.eigval
Threshold to remove "insignificant" singular vectors. Default is
1e-4.- ncores
Number of cores used. Default doesn't use parallelism. You may use nb_cores.
Value
A data.frame with 3 elements:
the slopes of each regression,
the standard errors of each slope,
the t-scores associated with each slope. This is also an object of class
mhtest. Seemethods(class = "mhtest").
See also
Examples
set.seed(1)
X <- big_attachExtdata()
n <- nrow(X)
y <- rnorm(n)
covar <- matrix(rnorm(n * 3), n)
X1 <- X[, 1] # only first column of the Filebacked Big Matrix
# Without covar
test <- big_univLinReg(X, y)
## New class `mhtest`
class(test)
#> [1] "mhtest" "data.frame"
attr(test, "transfo")
#> function (x) .Primitive("abs")
attr(test, "predict")
#> function (xtr)
#> {
#> lpval <- stats::pt(xtr, df = 515, lower.tail = FALSE, log.p = TRUE)
#> (log(2) + lpval)/log(10)
#> }
#> <environment: base>
## plot results
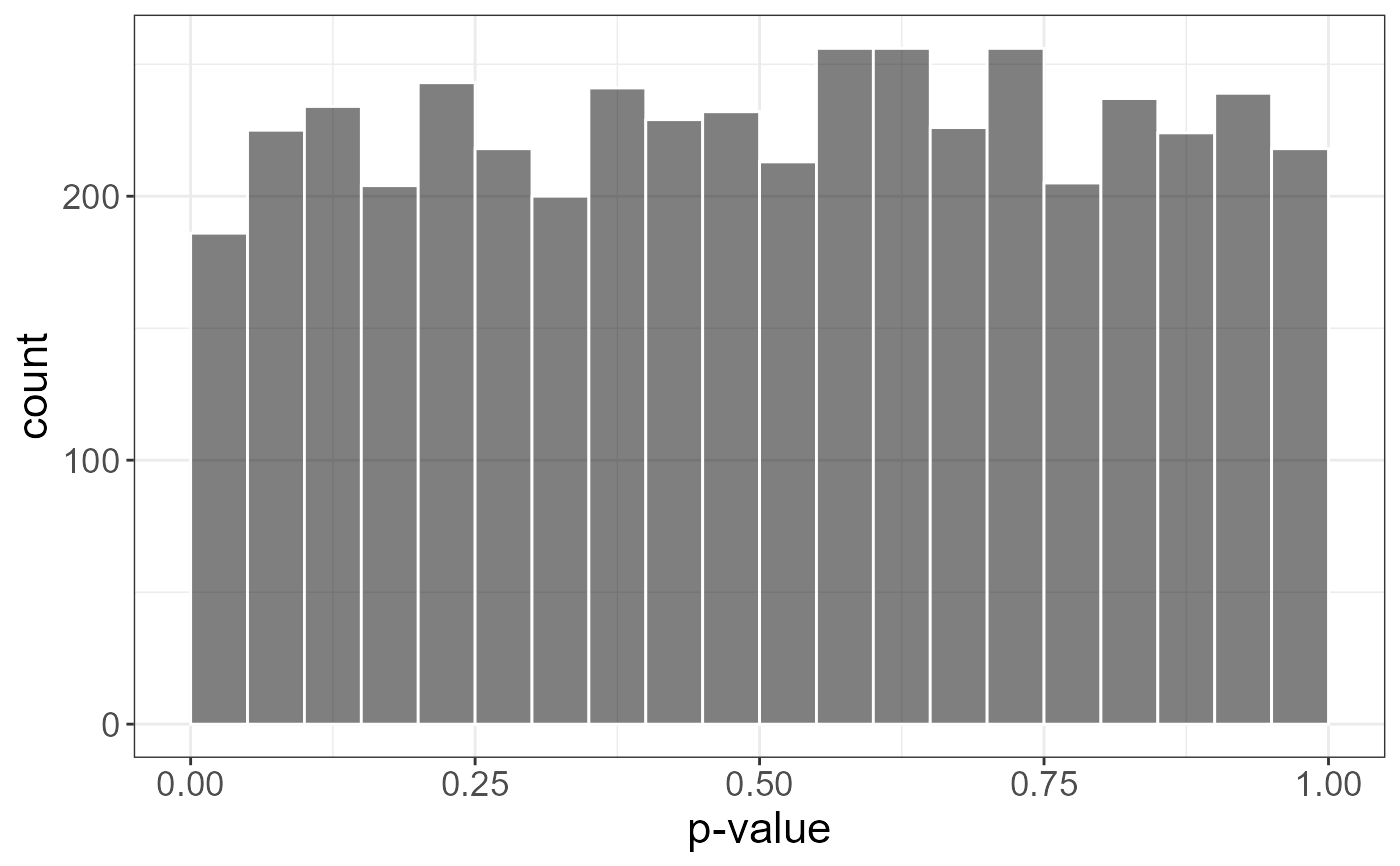
plot(test)
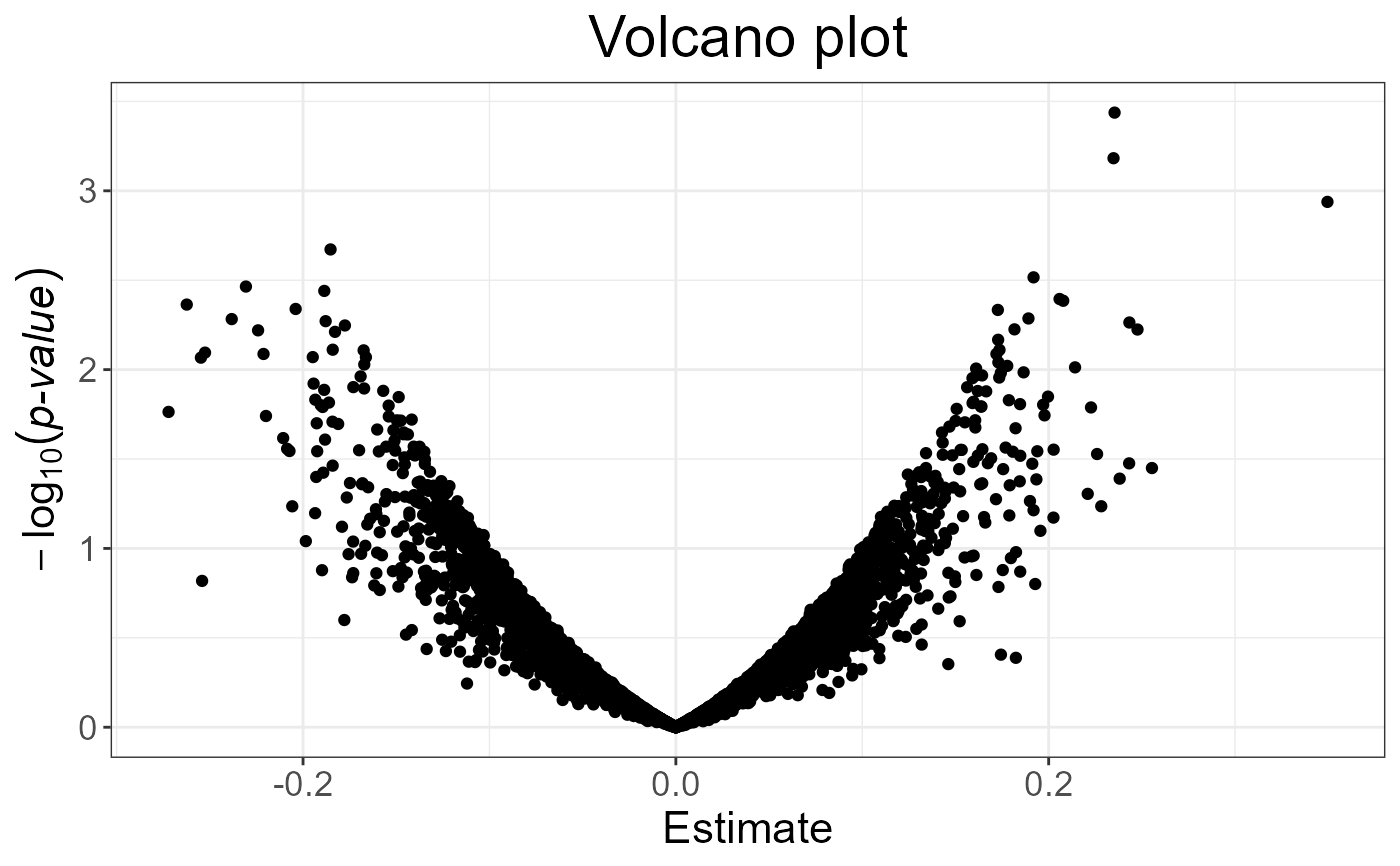
 plot(test, type = "Volcano")
plot(test, type = "Volcano")
 ## To get p-values associated with the test
test$p.value <- predict(test, log10 = FALSE)
str(test)
#> Classes 'mhtest' and 'data.frame': 4542 obs. of 4 variables:
#> $ estim : num -0.0311 0.0956 -0.0961 0.0327 -0.0246 ...
#> $ std.err: num 0.0658 0.0783 0.071 0.08 0.062 ...
#> $ score : num -0.473 1.221 -1.353 0.408 -0.397 ...
#> $ p.value: num 0.636 0.223 0.177 0.683 0.692 ...
#> - attr(*, "transfo")=function (x)
#> - attr(*, "predict")=function (xtr)
summary(lm(y ~ X1))$coefficients[2, ]
#> Estimate Std. Error t value Pr(>|t|)
#> -0.03111113 0.06575485 -0.47313823 0.63631503
# With all data
str(big_univLinReg(X, y, covar = covar))
#> Classes 'mhtest' and 'data.frame': 4542 obs. of 3 variables:
#> $ estim : num -0.0295 0.0994 -0.0939 0.035 -0.0217 ...
#> $ std.err: num 0.066 0.0785 0.0714 0.0803 0.0625 ...
#> $ score : num -0.447 1.266 -1.316 0.436 -0.347 ...
#> - attr(*, "transfo")=function (x)
#> - attr(*, "predict")=function (xtr)
summary(lm(y ~ X1 + covar))$coefficients[2, ]
#> Estimate Std. Error t value Pr(>|t|)
#> -0.02947921 0.06597253 -0.44684070 0.65517903
# With only half of the data
ind.train <- sort(sample(n, n/2))
str(big_univLinReg(X, y[ind.train],
covar.train = covar[ind.train, ],
ind.train = ind.train))
#> Classes 'mhtest' and 'data.frame': 4542 obs. of 3 variables:
#> $ estim : num 0.00416 0.15009 0.03116 0.16321 -0.02771 ...
#> $ std.err: num 0.0962 0.1131 0.1034 0.1102 0.0897 ...
#> $ score : num 0.0432 1.3267 0.3012 1.4807 -0.3088 ...
#> - attr(*, "transfo")=function (x)
#> - attr(*, "predict")=function (xtr)
summary(lm(y ~ X1 + covar, subset = ind.train))$coefficients[2, ]
#> Estimate Std. Error t value Pr(>|t|)
#> 0.004156134 0.096202616 0.043201885 0.965574678
## To get p-values associated with the test
test$p.value <- predict(test, log10 = FALSE)
str(test)
#> Classes 'mhtest' and 'data.frame': 4542 obs. of 4 variables:
#> $ estim : num -0.0311 0.0956 -0.0961 0.0327 -0.0246 ...
#> $ std.err: num 0.0658 0.0783 0.071 0.08 0.062 ...
#> $ score : num -0.473 1.221 -1.353 0.408 -0.397 ...
#> $ p.value: num 0.636 0.223 0.177 0.683 0.692 ...
#> - attr(*, "transfo")=function (x)
#> - attr(*, "predict")=function (xtr)
summary(lm(y ~ X1))$coefficients[2, ]
#> Estimate Std. Error t value Pr(>|t|)
#> -0.03111113 0.06575485 -0.47313823 0.63631503
# With all data
str(big_univLinReg(X, y, covar = covar))
#> Classes 'mhtest' and 'data.frame': 4542 obs. of 3 variables:
#> $ estim : num -0.0295 0.0994 -0.0939 0.035 -0.0217 ...
#> $ std.err: num 0.066 0.0785 0.0714 0.0803 0.0625 ...
#> $ score : num -0.447 1.266 -1.316 0.436 -0.347 ...
#> - attr(*, "transfo")=function (x)
#> - attr(*, "predict")=function (xtr)
summary(lm(y ~ X1 + covar))$coefficients[2, ]
#> Estimate Std. Error t value Pr(>|t|)
#> -0.02947921 0.06597253 -0.44684070 0.65517903
# With only half of the data
ind.train <- sort(sample(n, n/2))
str(big_univLinReg(X, y[ind.train],
covar.train = covar[ind.train, ],
ind.train = ind.train))
#> Classes 'mhtest' and 'data.frame': 4542 obs. of 3 variables:
#> $ estim : num 0.00416 0.15009 0.03116 0.16321 -0.02771 ...
#> $ std.err: num 0.0962 0.1131 0.1034 0.1102 0.0897 ...
#> $ score : num 0.0432 1.3267 0.3012 1.4807 -0.3088 ...
#> - attr(*, "transfo")=function (x)
#> - attr(*, "predict")=function (xtr)
summary(lm(y ~ X1 + covar, subset = ind.train))$coefficients[2, ]
#> Estimate Std. Error t value Pr(>|t|)
#> 0.004156134 0.096202616 0.043201885 0.965574678