LOF: Identifying Density-Based Local Outliers.
LOF(
U,
seq_k = c(4, 10, 30),
combine = max,
robMaha = FALSE,
log = TRUE,
ncores = 1
)Arguments
- U
A matrix, from which to detect outliers (rows). E.g. PC scores.
- seq_k
Sequence of numbers of nearest neighbors to use. If multiple
kare provided, this returns the combination of statistics. Default isc(4, 10, 30)and usemaxto combine (seecombine).- combine
How to combine results for multiple
k? Default usesmax.- robMaha
Whether to use a robust Mahalanobis distance instead of the normal euclidean distance? Default is
FALSE, meaning using euclidean.- log
Whether to return the logarithm of LOFs? Default is
TRUE.- ncores
Number of cores to use. Default is
1.
References
Breunig, Markus M., et al. "LOF: identifying density-based local outliers." ACM sigmod record. Vol. 29. No. 2. ACM, 2000.
See also
Examples
X <- readRDS(system.file("testdata", "three-pops.rds", package = "bigutilsr"))
svd <- svds(scale(X), k = 10)
llof <- LOF(svd$u)
hist(llof, breaks = nclass.scottRob)
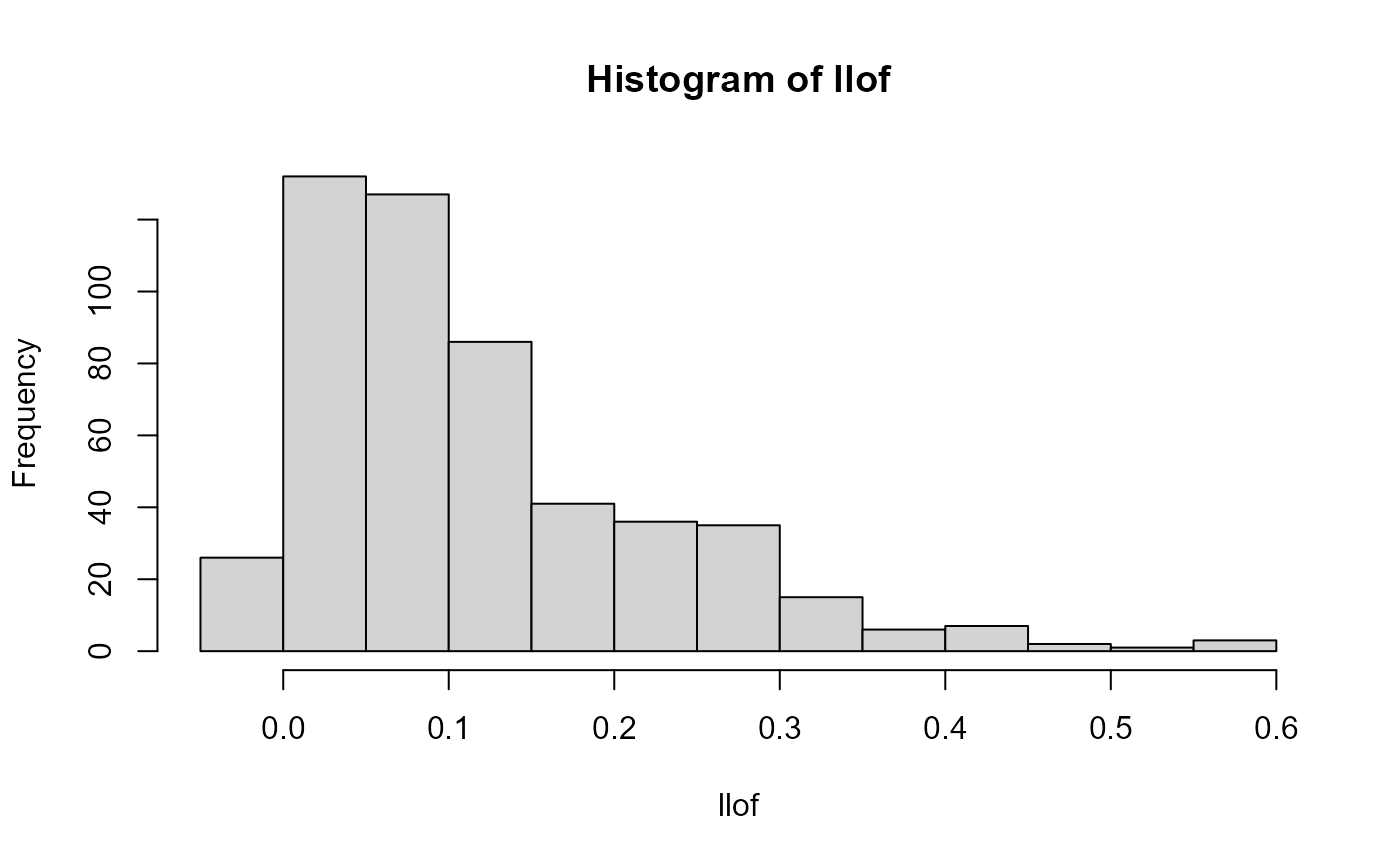 tukey_mc_up(llof)
#> [1] 0.8646255
llof_maha <- LOF(svd$u, robMaha = TRUE)
hist(llof_maha, breaks = nclass.scottRob)
tukey_mc_up(llof)
#> [1] 0.8646255
llof_maha <- LOF(svd$u, robMaha = TRUE)
hist(llof_maha, breaks = nclass.scottRob)
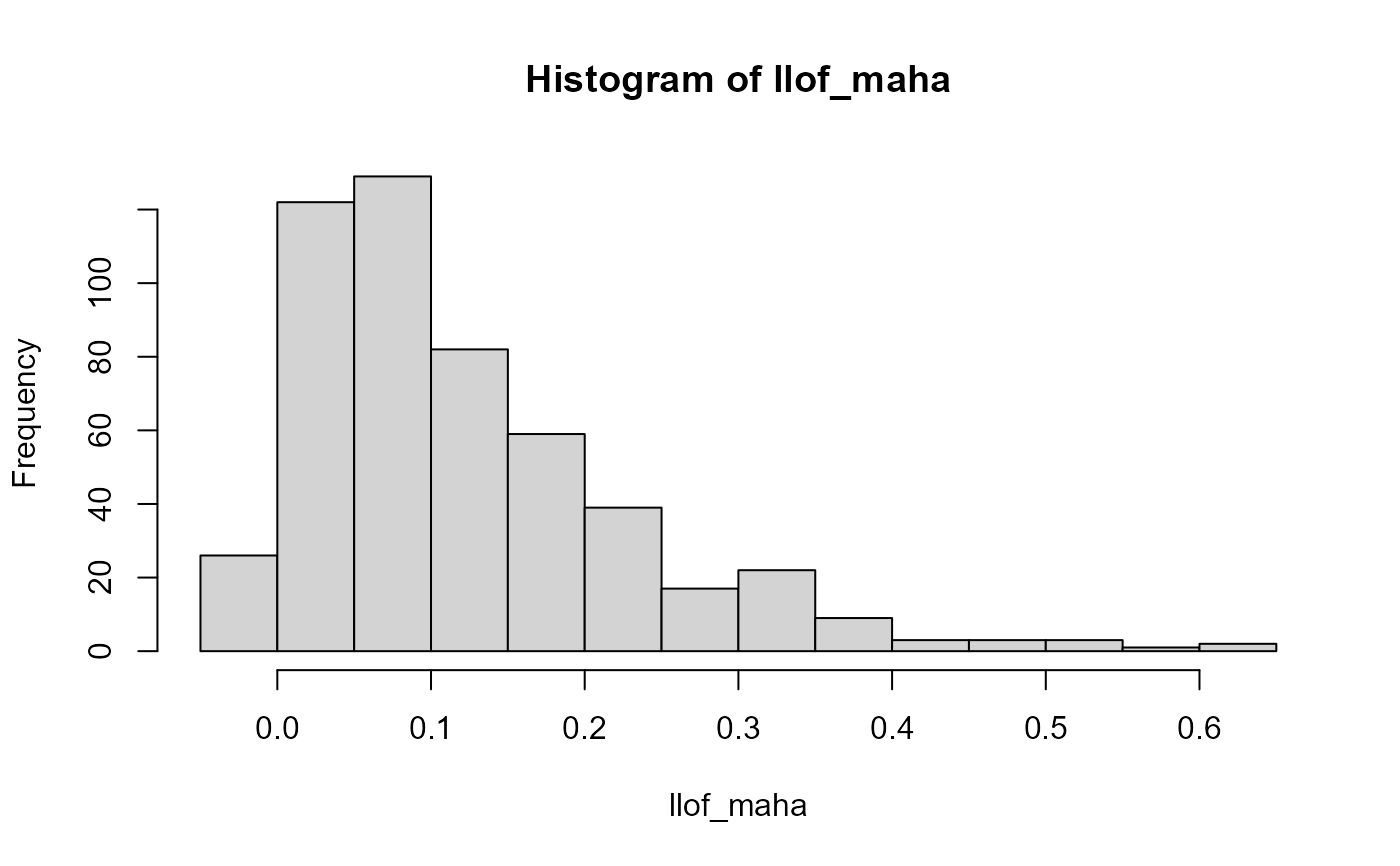 tukey_mc_up(llof_maha)
#> [1] 0.7319994
lof <- LOF(svd$u, log = FALSE)
hist(lof, breaks = nclass.scottRob)
tukey_mc_up(llof_maha)
#> [1] 0.7319994
lof <- LOF(svd$u, log = FALSE)
hist(lof, breaks = nclass.scottRob)
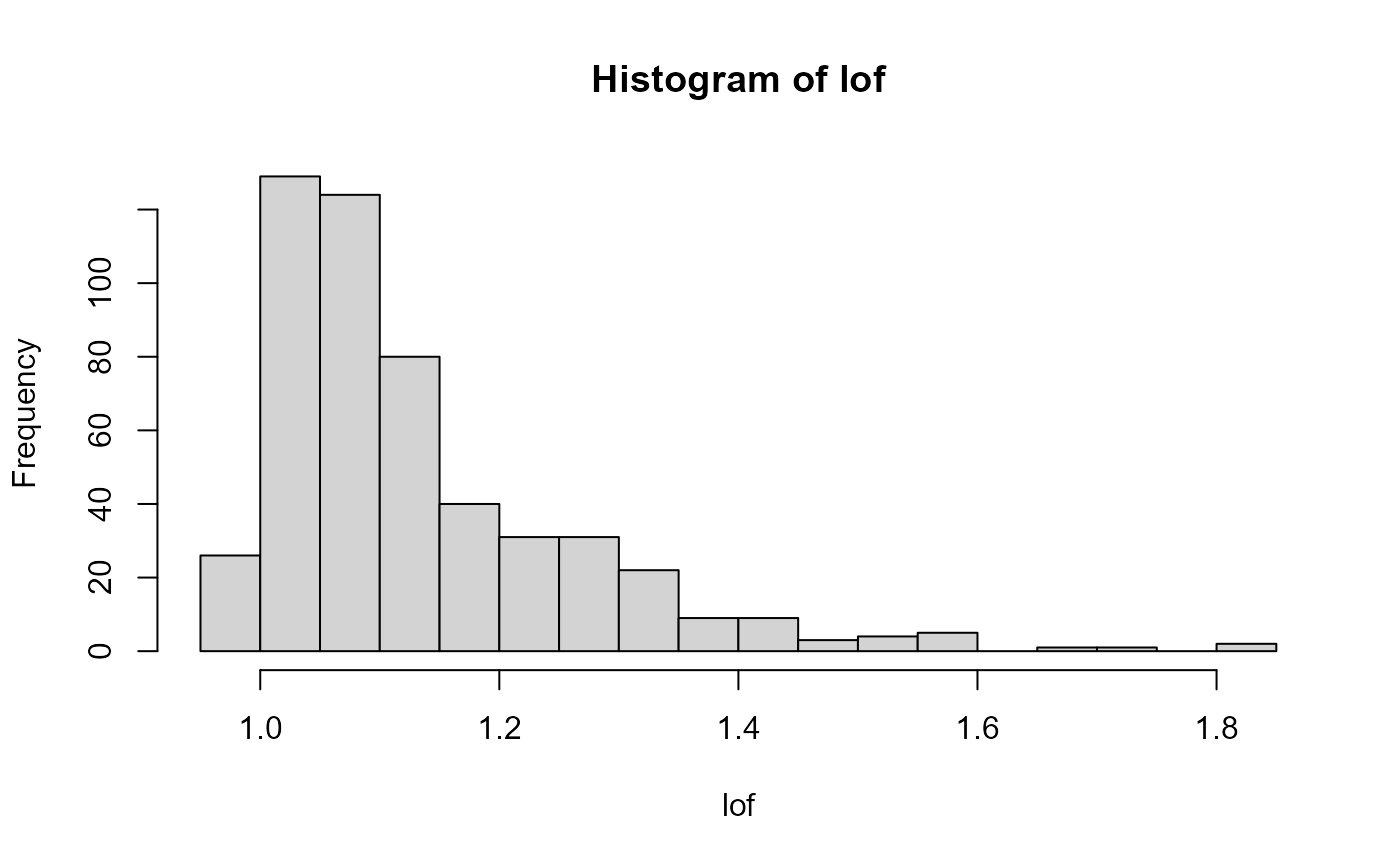 str(hist_out(lof))
#> List of 2
#> $ x : num [1:513] 1.04 1.15 1.08 1 1.01 ...
#> $ lim: num [1:2] -Inf 1.63
str(hist_out(lof, nboot = 100))
#> List of 3
#> $ x : num [1:513] 1.04 1.15 1.08 1 1.01 ...
#> $ lim : num [1:2] -Inf 1.63
#> $ all_lim: num [1:2, 1:100] -Inf 1.63 -Inf 1.63 -Inf ...
str(hist_out(lof, nboot = 100, breaks = "FD"))
#> List of 3
#> $ x : num [1:513] 1.04 1.15 1.08 1 1.01 ...
#> $ lim : num [1:2] -Inf 1.63
#> $ all_lim: num [1:2, 1:100] -Inf 1.63 -Inf 1.63 -Inf ...
str(hist_out(lof))
#> List of 2
#> $ x : num [1:513] 1.04 1.15 1.08 1 1.01 ...
#> $ lim: num [1:2] -Inf 1.63
str(hist_out(lof, nboot = 100))
#> List of 3
#> $ x : num [1:513] 1.04 1.15 1.08 1 1.01 ...
#> $ lim : num [1:2] -Inf 1.63
#> $ all_lim: num [1:2, 1:100] -Inf 1.63 -Inf 1.63 -Inf ...
str(hist_out(lof, nboot = 100, breaks = "FD"))
#> List of 3
#> $ x : num [1:513] 1.04 1.15 1.08 1 1.01 ...
#> $ lim : num [1:2] -Inf 1.63
#> $ all_lim: num [1:2, 1:100] -Inf 1.63 -Inf 1.63 -Inf ...