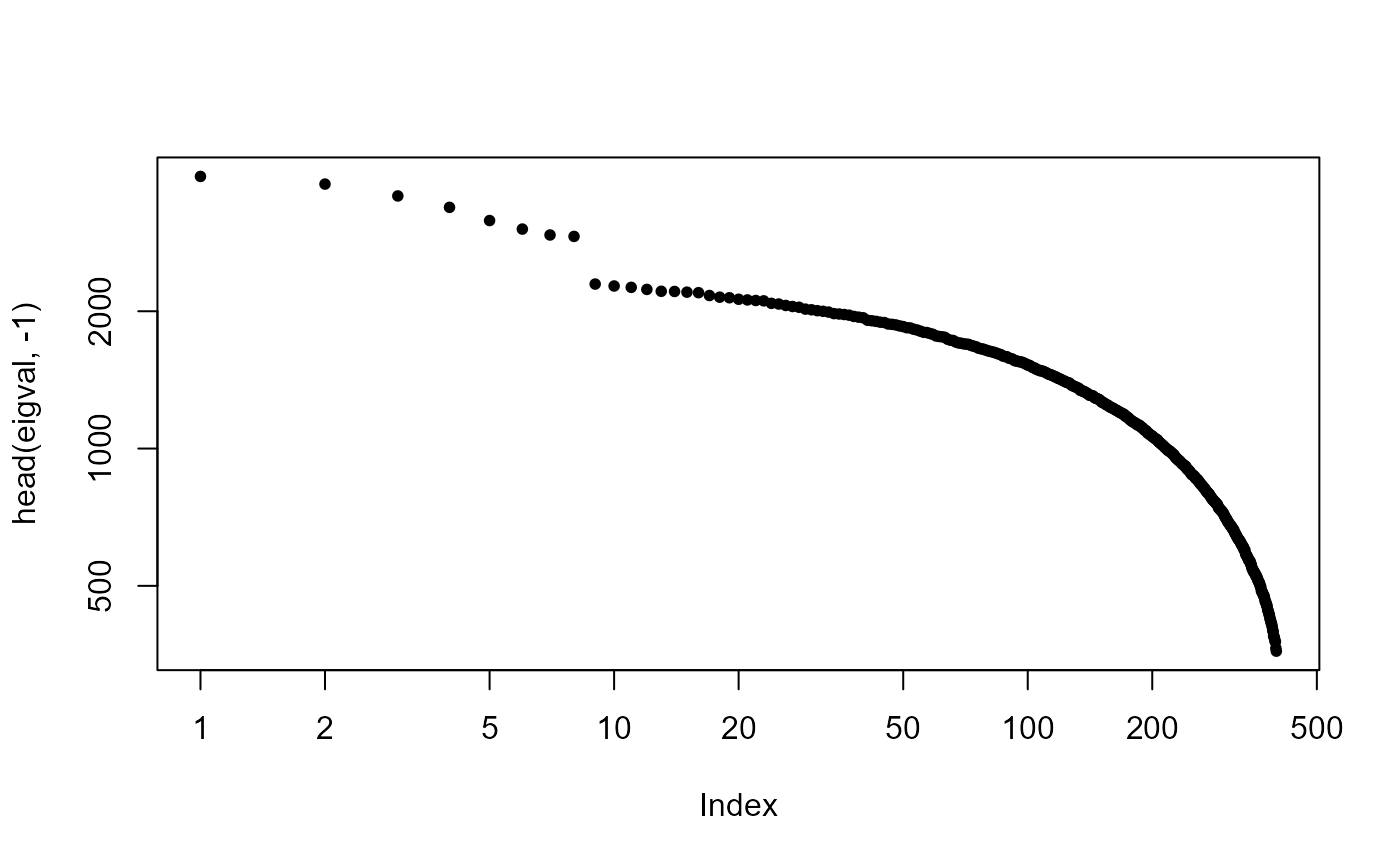
Estimate the number of distant spikes based on the histogram of eigenvalues.
pca_nspike(eigval, breaks = "FD", nboot = 100)Arguments
- eigval
Eigenvalues (squared singular values).
- breaks
Same parameter as for
hist(). Default uses a robust version of Scott's rule. You can also use"FD"ornclass.FDfor a bit more bins.- nboot
Number of bootstrap replicates to estimate limits more robustly. Default is
100.
Value
The estimated number of distant spikes.