Procrustes transform Y = pXR (after centering), where p is a scaling coefficient and R is a rotation matrix that minimize ||Y - pXR||_F.
procrustes(Y, X, n_iter_max = 1000, epsilon_min = 1e-07)Arguments
- Y
Reference matrix.
- X
Matrix to transform (
ncol(X) >= ncol(Y)).- n_iter_max
Maximum number of iterations. Default is
1000.- epsilon_min
Convergence criterion. Default is
1e-7.
Value
Object of class "procrustes", a list with the following elements:
$R: the rotation matrix to apply toX,$rho: the scaling coefficient to apply toX,$c: the column centering to apply to the resulting matrix,$diff: the average difference betweenYandXtransformed.
You can use method predict() to apply this transformation to other data.
Examples
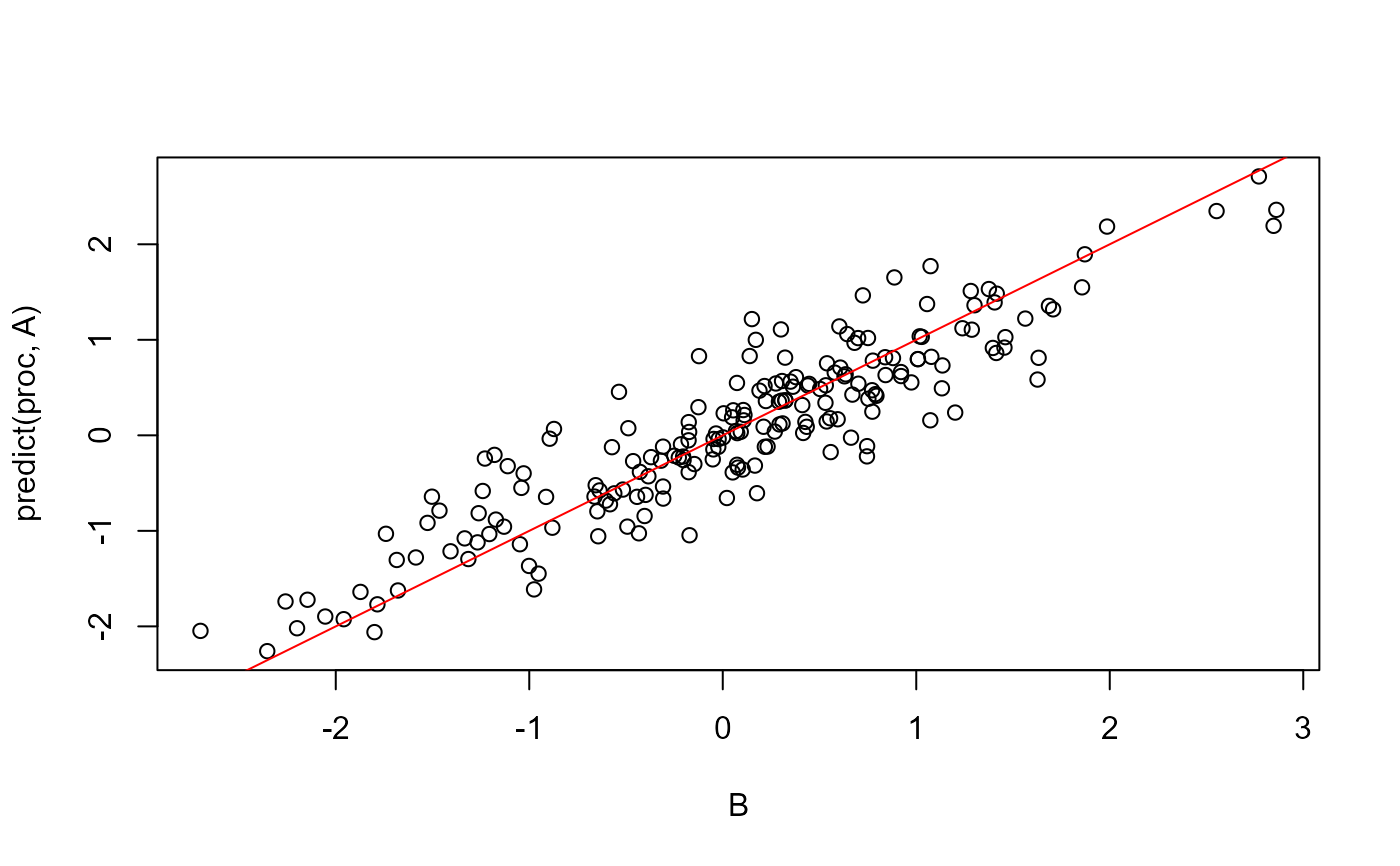
A <- matrix(rnorm(200), ncol = 20)
B <- matrix(rnorm(length(A)), nrow = nrow(A))
proc <- procrustes(B, A)
str(proc)
#> List of 4
#> $ R : num [1:20, 1:20] 0.3607 -0.3925 0.2973 -0.0806 0.0411 ...
#> $ rho : num 0.942
#> $ c : num [1, 1:20] 0.184 0.422 0.555 0.352 -0.107 ...
#> $ diff: num 0.177
#> - attr(*, "class")= chr "Procrustes"
plot(B, predict(proc, A)); abline(0, 1, col = "red")