Using clustering to find points in an image
In this post, I present my new package {img2coord}. This package can be used to retrieve coordinates from a scatter plot (as an image).
devtools::install_github("privefl/img2coord")Have you ever made a plot, saved it as a png and moved on? When you come back to it, it is sometimes difficult to read the values from this plot, especially if there is no grid inside the plot.
Making this package was also a good way to practice with clustering.
A very simple example
Saving a plot as PNG
file <- tempfile(fileext = ".png")
png(file, width = 600, height = 400)
set.seed(1)
plot(c(0, runif(20), 1))
dev.off()## png
## 2Reading the PNG in R
(img <- magick::image_read(file))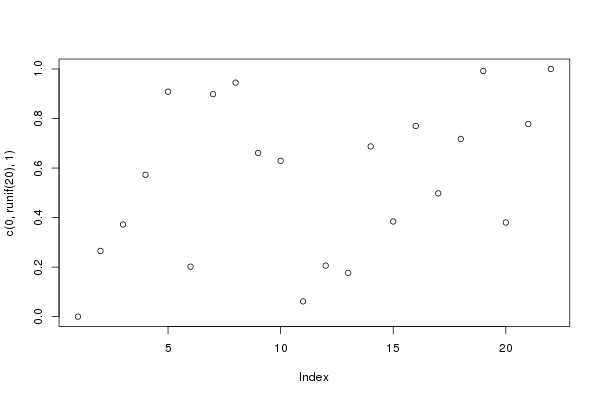
Get pixel indices from points
## grayscale
img_mat <- img2coord:::img2mat(img)
dim(img_mat)## [1] 400 600list.contour <- img2coord:::get_contours(img_mat)
img_mat_in <- img2coord:::get_inside(img_mat, list.contour)
dim(img_mat_in)## [1] 264 507head(ind <- which(img_mat_in > 0, arr.ind = TRUE))## row col
## [1,] 256 14
## [2,] 257 14
## [3,] 254 15
## [4,] 255 15
## [5,] 256 15
## [6,] 257 15Cluster pixel indices
set.seed(1)
km <- kmeans(ind, centers = 22)library(ggplot2)
myplot <- function(points, centers) {
p <- ggplot() +
geom_tile(aes(col, row), data = as.data.frame(points)) +
geom_point(aes(col, row), data = as.data.frame(centers), col = "red") +
bigstatsr::theme_bigstatsr() +
coord_equal()
print(p)
}
myplot(ind, km$centers)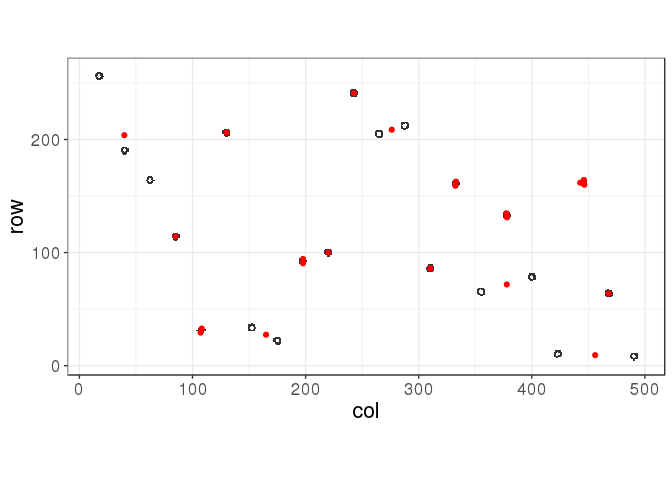
Even when using the true number of clusters, kmeans get trapped in a local minimum (this is clearly not the best solution!), depending on the initialisation of centers. One possible solution would be to use many initialisations; let’s try that.
set.seed(1)
km <- kmeans(ind, centers = 22, nstart = 100, iter.max = 100)## Warning: did not converge in 100 iterations
## Warning: did not converge in 100 iterationsmyplot(ind, km$centers)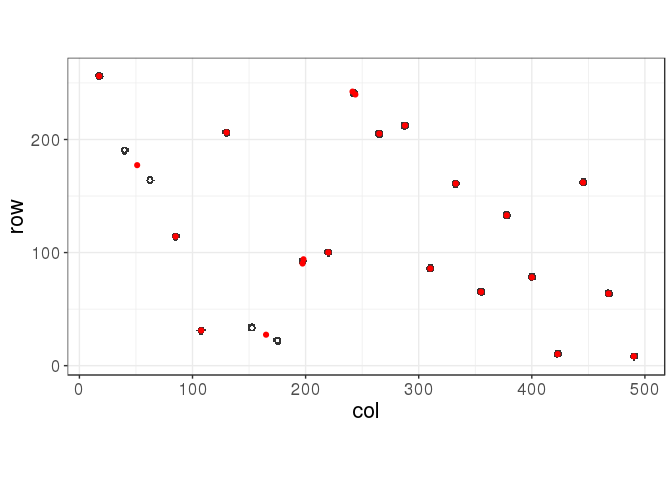
It is better but not optimal.
Using hclust to get centers
get_centers <- function(points, clusters) {
do.call("rbind", by(points, clusters, colMeans, simplify = FALSE))
}
d <- dist(ind)
hc <- hclust(d)
centers <- get_centers(ind, cutree(hc, k = 22))
myplot(ind, centers)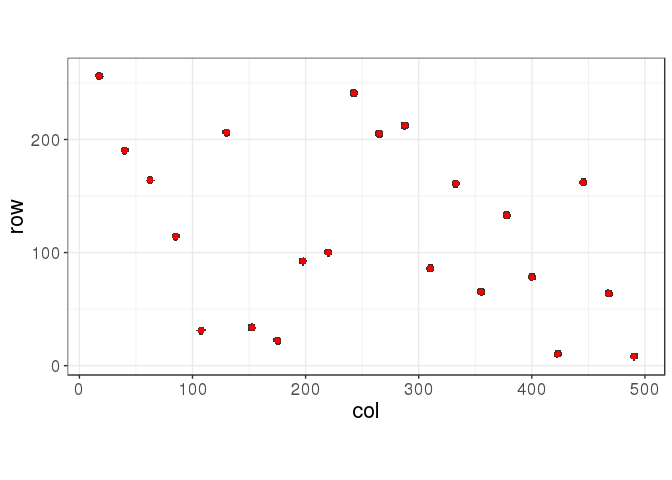
hclust() works well for this example.
Get the number of clusters
What if we don’t know the number of clusters (representing the initial points)? A statistic that could help us determine the number of clusters to use is the silhouette.
K_seq <- seq(10, 30)
stat <- sapply(K_seq, function(k) {
mean(cluster::silhouette(cutree(hc, k), d)[, 3])
})
plot(K_seq, stat, pch = 20); abline(v = 22, lty = 3)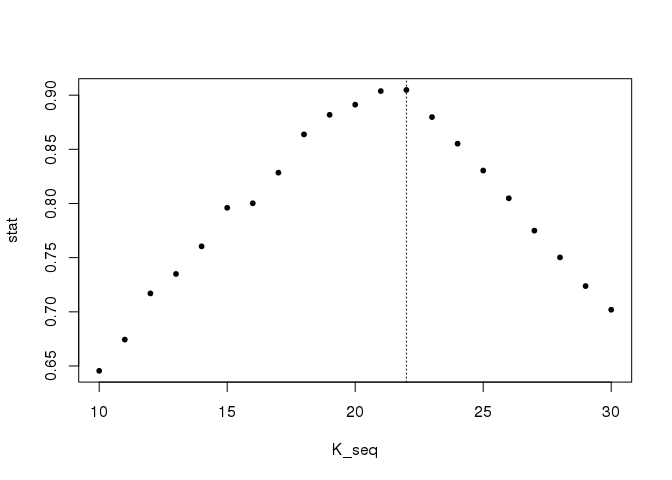
A less simple example
file <- tempfile(fileext = ".png")
png(file, width = 600, height = 400)
set.seed(1)
y <- c(0, runif(100), 1)
plot(y, cex = runif(102, min = 0.5, max = 1.5))
dev.off()## png
## 2(img <- magick::image_read(file))
img_mat <- img2coord:::img2mat(img)
list.contour <- img2coord:::get_contours(img_mat)
img_mat_in <- img2coord:::get_inside(img_mat, list.contour)
ind <- which(img_mat_in > 0, arr.ind = TRUE)hc <- flashClust::hclust(d <- dist(ind))
K_seq <- seq(50, 150)
stat <- sapply(K_seq, function(k) {
mean(cluster::silhouette(cutree(hc, k), d)[, 3])
})
plot(K_seq, stat, pch = 20); abline(v = 102, lty = 3)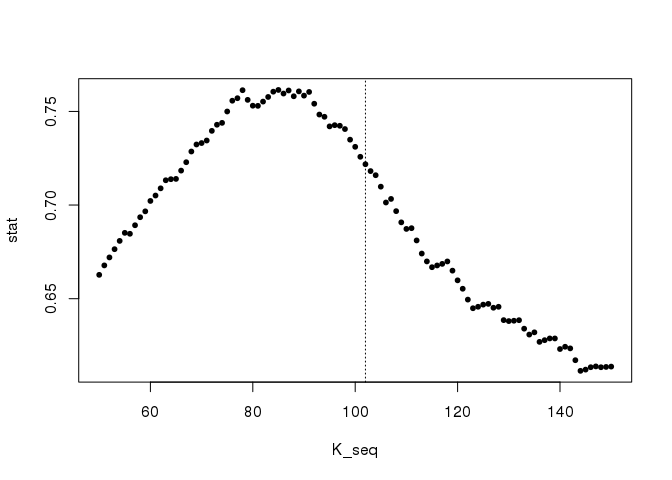
(K_opt <- K_seq[which.max(stat)])## [1] 85centers <- get_centers(ind, cutree(hc, k = K_opt))
myplot(ind, centers)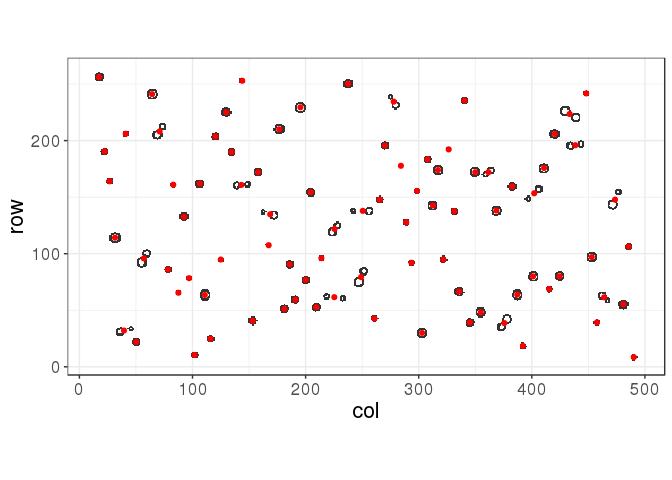
The silhouette statistic is giving a good yet not optimal solution in this situation. Using the true number of points, we would get:
centers <- get_centers(ind, cutree(hc, k = 102))
myplot(ind, centers)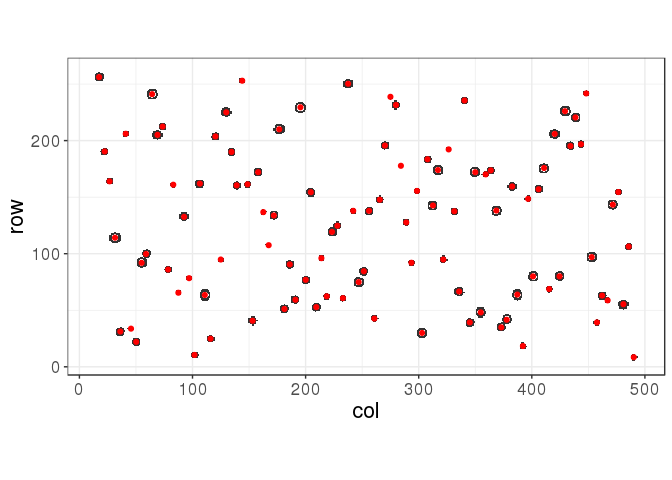
If someone has a better statistic to (automatically) find the number of clusters, please share it and I’ll update this post.
Putting everything together as a package
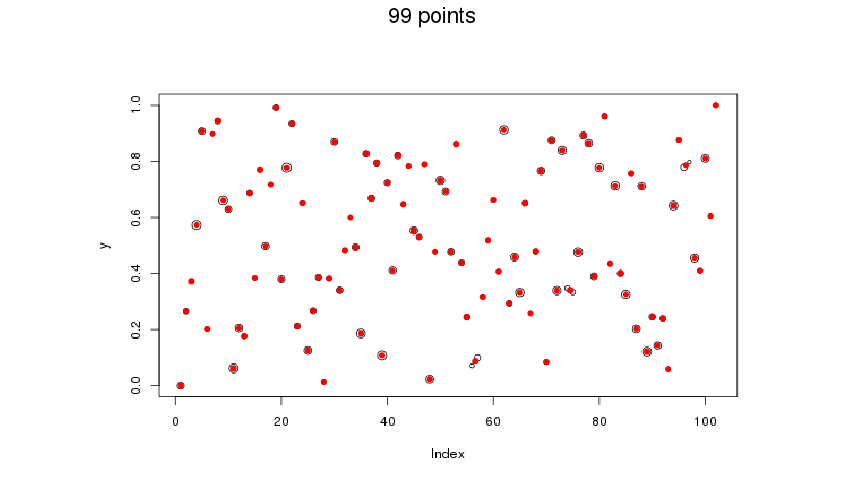
Finally, after you get the center of all points (pixel clusters), you can interpolate the values based on the values of axe ticks.
coord <- img2coord::get_coord(
file,
x_ticks = seq(0, 100, 20),
y_ticks = seq(0, 1, 0.2),
K_min = 50, K_max = 150
) 
This works better here because I combined the silhouette statistic with a gini coefficient (measure of dispersion) of the number of pixels in each cluster (assuming that they should have approximately the same number). Let’s have a look at the combined statistic:
stat <- attr(coord, "stat")
plot(names(stat), stat, pch = 20); abline(v = 102, lty = 3) 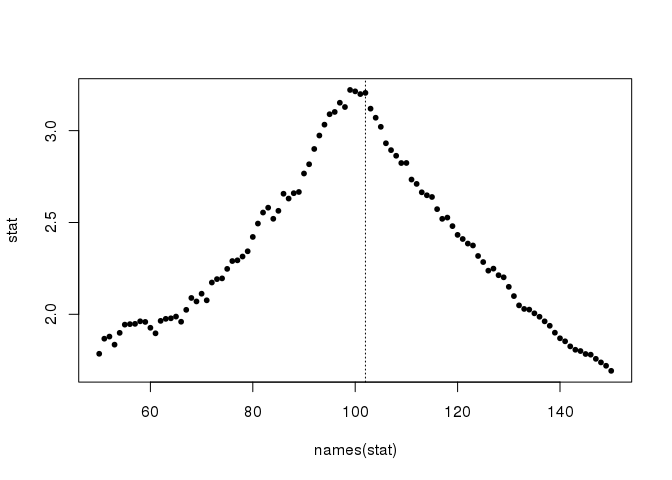
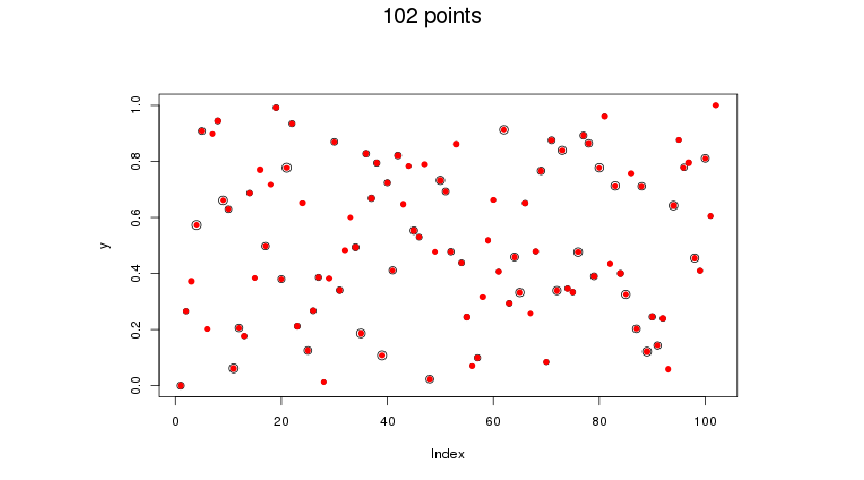
If you don’t get the right number of clusters the first time, you can use the plot generated by img2coord::get_coord() to adjust K.
coord <- img2coord::get_coord(
file,
x_ticks = seq(0, 100, 20),
y_ticks = seq(0, 1, 0.2),
K = 102 ## 99 + 3
) 
Let’s verify the coordinates we get:
round(coord$x, 2)## [1] 1.00 2.01 3.01 4.00 4.99 6.01 7.00 8.00 9.01 10.01
## [11] 10.98 11.99 12.99 14.01 15.00 15.98 17.01 18.00 18.98 20.00
## [21] 20.99 22.00 23.00 23.99 24.98 26.00 26.99 28.01 29.00 29.98
## [31] 31.01 31.99 33.02 34.00 35.02 35.98 37.00 38.01 38.99 40.00
## [41] 40.99 41.99 42.99 44.01 44.97 45.98 47.01 48.00 49.00 50.00
## [51] 51.01 51.98 52.99 53.99 55.00 56.00 57.01 58.01 59.00 60.02
## [61] 61.00 62.00 62.98 64.01 64.99 65.98 67.00 68.01 68.98 70.00
## [71] 70.98 72.00 73.00 74.01 75.00 76.01 77.00 78.00 79.02 79.98
## [81] 81.00 82.01 82.98 84.00 85.01 85.99 87.01 87.98 88.99 89.99
## [91] 90.98 92.00 93.00 94.01 95.00 95.89 96.87 97.99 99.00 100.00
## [101] 101.01 101.99plot(coord$y, y, pch = 20); abline(0, 1, col = "red")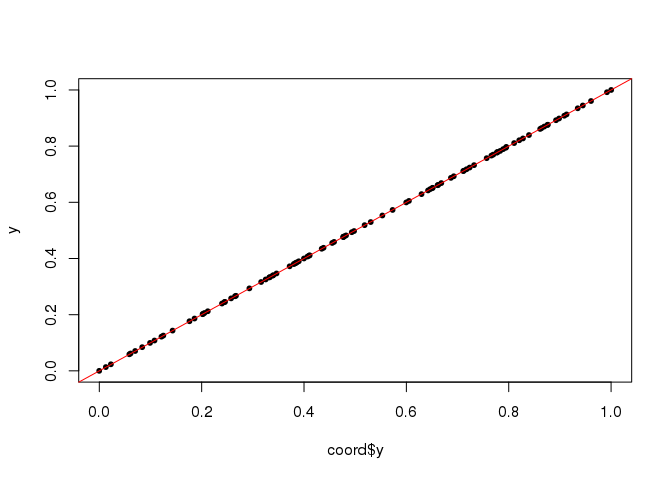
Handling large images
url <- "https://goo.gl/K6Y7D1"
library(img2coord)
(img <- img_read(url))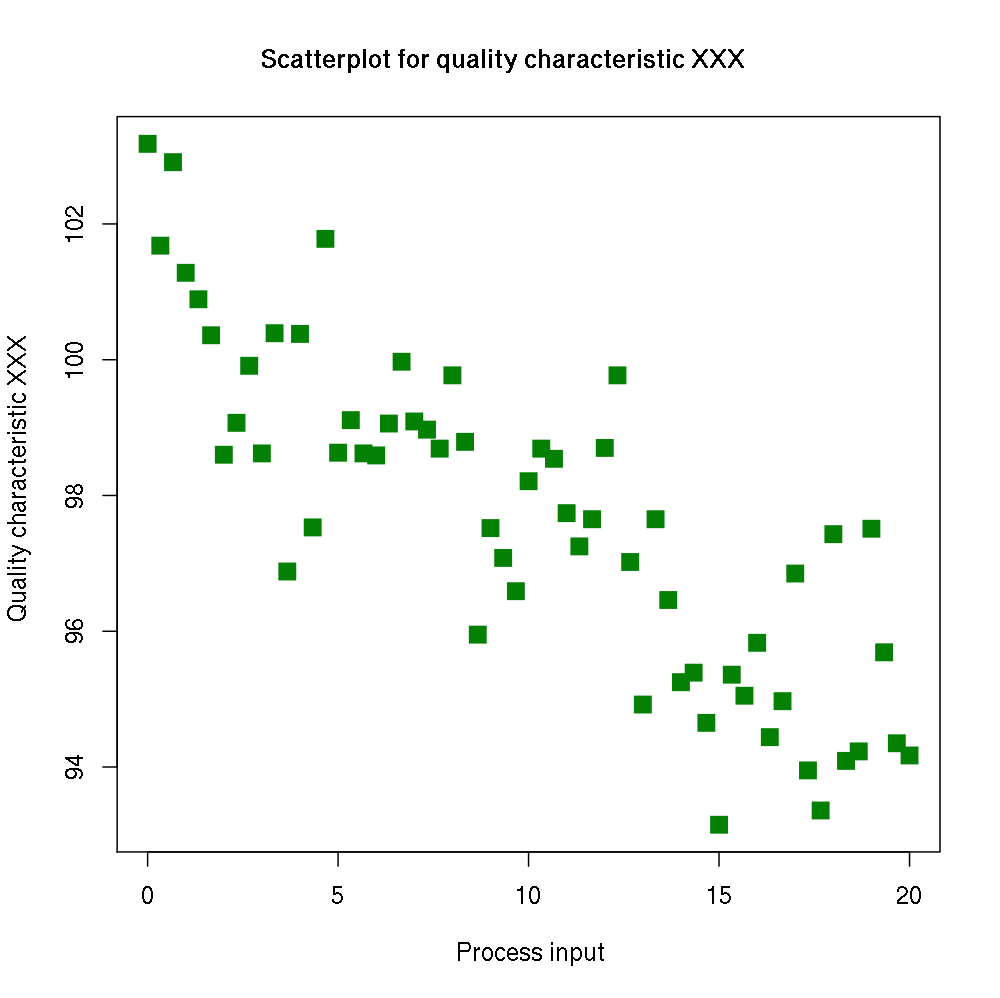
get_coord(img, seq(0, 20, 5), seq(94, 102, 2), K_min = 40, K_max = 80)## Error: Detected more than 10000 pixels associated with points (21358).
## Make sure you have a white background with no grid (only points).
## You can change 'max_pixels', but it could become time/memory consuming.
## You can also downsize the image using `img_scale()`.The green points are spanning 21,358 pixels, which could be a lot to process, depending on your computer. To solve this problem, you can do:
img %>%
img_scale(0.4) %>%
get_coord(seq(0, 20, 5), seq(94, 102, 2), K_min = 40, K_max = 80)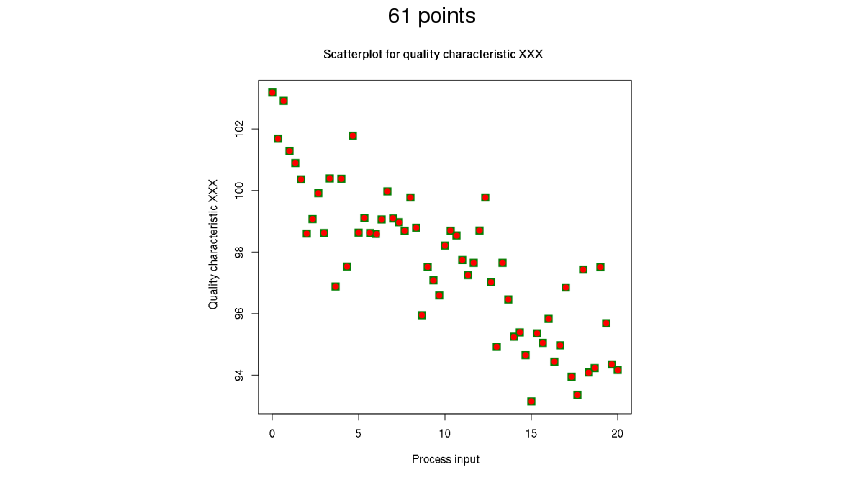
## $x
## [1] -0.0005401687 0.3343468897 0.6664667098 0.9992303467 1.3337481120
## [6] 1.6671560578 1.9997172265 2.3332269701 2.6674887594 2.9991939098
## [11] 3.3326859175 3.6672015650 4.0000566377 4.3329094030 4.6674459579
## [16] 5.0009192485 5.3326185095 5.6478368286 6.0464367945 6.3329501844
## [21] 6.6663573828 6.9839799351 7.3513525635 7.6674682785 8.0006621331
## [26] 8.3329974666 8.6657575234 8.9999189758 9.3339407034 9.6666009658
## [31] 10.0001231324 10.3173252500 10.6778169496 10.9989528978 11.3335964907
## [36] 11.6665738876 11.9994145980 12.3336165187 12.6676553366 12.9992455320
## [41] 13.3331964821 13.6668686496 13.9813979399 14.3425396058 14.6672323938
## [46] 15.0002253318 15.3330976683 15.6669750015 16.0007098735 16.3322818494
## [51] 16.6663301257 17.0005035442 17.3333809139 17.6663416006 18.0009680691
## [56] 18.3172429708 18.6798897677 18.9998067488 19.3333374006 19.6570288544
## [61] 20.0116006657
##
## $y
## [1] 103.18007 101.68089 102.91175 101.28109 100.89144 100.36108 98.59931
## [8] 99.06933 99.90900 98.61988 100.39017 96.87960 100.38054 97.52969
## [15] 101.77948 98.63014 99.10885 98.62457 98.58221 99.06001 99.97088
## [22] 99.09976 98.96014 98.68939 99.77109 98.78963 95.94976 97.51865
## [29] 97.07733 96.58967 98.21105 98.69922 98.53481 97.74167 97.24962
## [36] 97.65023 98.70168 99.77109 97.02095 94.91963 97.65023 96.46068
## [43] 95.23798 95.39595 94.64729 93.14815 95.35968 95.04774 95.82991
## [50] 94.43923 94.96829 96.84944 93.94962 93.35852 97.42928 94.07914
## [57] 94.23708 97.51029 95.68894 94.35497 94.16229
##
## attr(,"stat")
## 40 41 42 43 44 45 46
## 1.991872 2.074327 2.140775 2.270267 2.341965 2.529008 2.758398
## 47 48 49 50 51 52 53
## 2.920247 3.119059 3.244979 3.377485 3.715905 4.083564 4.563417
## 54 55 56 57 58 59 60
## 5.104999 5.841044 6.441349 6.551265 7.246189 8.165916 8.842711
## 61 62 63 64 65 66 67
## 10.167292 8.357433 6.887889 5.708635 4.889674 4.274374 3.788661
## 68 69 70 71 72 73 74
## 3.384619 3.082882 2.836479 2.630589 2.451460 2.292444 2.162840
## 75 76 77 78 79 80
## 2.054934 1.947805 1.870104 1.784646 1.701550 1.630990Conclusion
We have seen that hclust() was performing better than kmeans() (for this example). For some reason I don’t understand yet, initializing kmeans() with centers from hclust() works even better.
Then, we have seen how to determine the number of clusters. Finally, we have seen that using a particular statistic, specifically designed for this problem, improved the solution.
Of course, this could be improved a lot. For example, this won’t work for plots having a background color or some grid inside. Feel free to bring your ideas. BTW, thanks Robin who brought some nice ideas that improved this package a lot.
Have a look at the GitHub repo.