Polygenic scores from individual-level data
Florian Privé
July 4, 2025
Source:vignettes/PLR.Rmd
PLR.RmdIf you have individual-level data (i.e. genotypes and phenotypes), you can basically use any supervised learning (machine learning) method to train a PGS. However, because of the size of the genetic data, you will quickly have scalability issues with these models. Moreover, it has been shown that effects for most diseases and traits are small and essentially additive, and that fancy methods such as deep learning are not much effective at constructing PGS (Kelemen et al. 2025).
Therefore, using penalized linear/logistic regression (PLR) can be a very efficient and effective method to train PGS. In my R package bigstatsr, I have developed a very fast implementation with automatic choice of the two hyper-parameters (Privé, Aschard, and Blum 2019). You can find a tutorial explaining its implementation and use here.
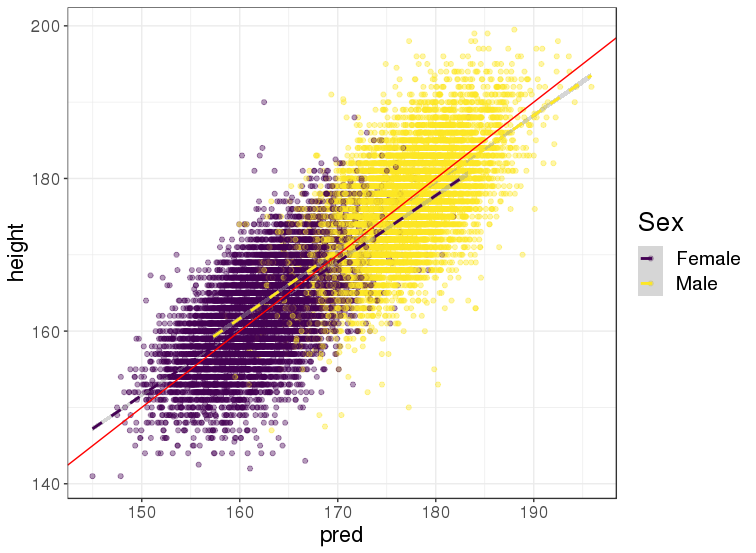
This is an example of using PLR for predicting height from genotypes in the UK Biobank
training on 350K individuals x 656K variants in less than 24H
within both males and females, PGS achieved a correlation of 65.5% (\(r^2\) of 42.9%) between genetically predicted and true height